Tutorial: Network representation in Teneto¶
There are three ways that network’s are represented in Teneto:
- A TemporalNetwork object
- Numpy array/snapshot
- Dictionary/contact representation
This tutorial goes through what these different representations. Teneto is migrating towards the TemporalNetwork object. However, it is possible to still use with the other two representations.
TemporalNetwork object¶
TemporalNetwork is a class in teneto.
>>> from teneto import TemporalNetwork
>>> tnet = TemporalNetwork()
...
As an input, you can pass it a 3D numpy array, a contact representation (see below), a list of edges or a pandas df (see below).
A feature of the TemporalNetwork class is that the different functions such as plotting and networkmeasures can be accessed within the object.
For example, the code below calls the function teneto.generatenetwork.rand_binomial with all subsequent arguments being arguments for the rand_binomial function:
>>> import numpy as np
>>> np.random.seed(2019) # Set random seed for replication
>>> tnet.generatenetwork('rand_binomial',size=(5,3), prob=0.5)
The data this creates is found in tnet.network which is a pandas data frame. To have a peak at the top of the data frame, we can call:
>>> tnet.network.head()
i j t
0 0 1 0
1 0 1 1
2 0 2 0
3 0 2 1
4 0 2 2
Each line in the data frame represents one edge. i and j are both node indexes and t is a temporal index. These column names are always present in data frames made by Teneto. There is no weight column here which indicates this is a binary network.
Exploring the network¶
You can inspect different parts of the network by calling tnet.get_network_when() and specifying an i, j or t argument.
>>> tnet.get_network_when(i=1)
i j t
6 1 2 0
7 1 2 2
8 1 3 0
9 1 4 1
The different argument can also be combined.
>>> tnet.get_network_when(i=1, t=0)
i j t
6 1 2 0
8 1 3 0
Lists can also be specified as arguments:
>>> tnet.get_network_when(i=[0, 1], t=1)
i j t
1 0 1 1
3 0 2 1
5 0 4 1
9 1 4 1
The logic within each argument is OR (i.e. about get all where i == 1 OR i == 0). The logic between the different arguments, defaults to AND. (i.e. get when i == [0 or 1] AND t == 1). In some cases, you may might the between argument logic to be OR:
>>> tnet.get_network_when(i=1, j=1, logic='or')
i j t
0 0 1 0
1 0 1 1
6 1 2 0
7 1 2 2
8 1 3 0
9 1 4 1
In the above case we select all edges where i == 1 OR j == 1.
Weighted networks¶
When a network is weighted, the weight appears in its own column in the pandas data frame.
>>> np.random.seed(2019) # For reproducibility
>>> G = np.random.beta(1, 1, [5,5,3]) # Creates 5 nodes and 3 time-points
>>> tnet = TemporalNetwork(from_array=G, nettype='wd', diagonal=True)
>>> tnet.network.head()
i j t weight
0 0 0 0 0.628820
1 0 0 1 0.059084
2 0 0 2 0.833974
3 0 1 0 0.856509
4 0 1 1 0.518670
Self edges get deleted unless the argument diagonal=True is passed. Above we can see that there are edges when both i and j are 0.
Dense and sparse networks¶
The example we saw previously was of a sparse network representation. This means that only the active connections are encoded in the representation and all other edges can be assumed to be zero/absent.
There are many weighted networks all edges have a value. These networks are called dense.
In denser networks, tnet.network will be a numpy array with node,node,time dimensions. The reason for this is simply speed. If you do not want a dense network to be created, you can pass a forcesparse=True argument when creating the TemporalNetwork.
If teneto is slow, it could be that creating the sparse network is taking too much time. So one way to ensure the dense representation is forced is to set the parameter dense_threshold. The default value is 0.1 (i.e. 10%), which means that if 10% of the network’s connections are present, teneto will make the network dense. But you can set this to any value.
The TemporalNetwork functions such as get_network_when() still function with the dense representation.
Exporting to a numpy array¶
You can export the network to a numpy array from the pandas data frame by calling to array:
>>> np.random.seed(2019) # For reproducibility
>>> G = np.random.beta(1, 1, [5,5,3]) # Creates 5 nodes and 3 time-points
>>> tnet = TemporalNetwork(from_array=G, nettype='wd', diagonal=True)
>>> G2 = tnet.to_array()
>>> G == G2
True
Here G2 is a 3D numpy array which is equal to the input G (a numpy array).
Meta-information¶
Within the object there are multiple bits of information about the network. We, for example, check that the above network create below is binary:
>>> tnet = TemporalNetwork()
>>> tnet.generatenetwork('rand_binomial',size=(3,5), prob=0.5)
>>> tnet.nettype
'bu'
- There are 4 different nettypes:
- bu, wu, wd and bd.
where b is for binary, w is for weighted, u means undirected and d means directed. Teneto tries to estimate the nettype, but specifying it is good practice.
You can also get the size of the network by using:
>>> tnet.netshape
(3, 5)
Which means there are 3 nodes and 5 time-points.
Certain metainformation is automatically used in the plotting tools. For example, you can add some meta information using the nodelabels (give names to the nodes), timelabels (give names to the time points), and timeunit arguments.
>>> import matplotlib.pyplot as plt
>>> tlabs = ['2014','2015','2016','2017','2018']
>>> tunit = 'years'
>>> nlabs = ['Ashley', 'Blake', 'Casey']
>>> tnet = TemporalNetwork(nodelabels=nlabs, timeunit=tunit, timelabels=tlabs, nettype='bu')
>>> tnet.generatenetwork('rand_binomial',size=(3,5), prob=0.5)
>>> tnet.plot('slice_plot', cmap='Set2')
>>> plt.show()
(Source code, png, hires.png, pdf)
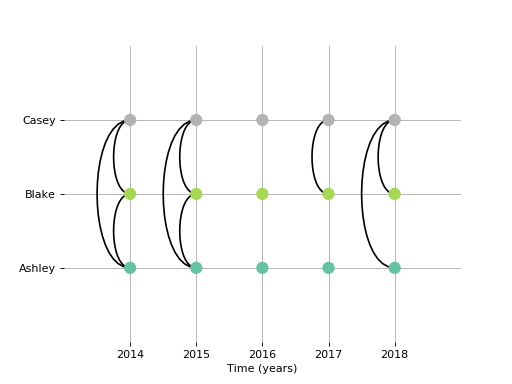
Importing data to TemporalNetwork¶
There are multiple ways to add data to the TemporalNetwork object. These include:
- A 3D numpy array
- Contact representation
- Pandas data frame
- List of edges
Numpy Arrays¶
For example, here we create a random network based on a beta distribution.
>>> np.random.seed(2019)
>>> G = np.random.beta(1, 1, [5,5,3])
>>> G.shape
(5, 5, 3)
Numpy arrays can get added by using the from_array argument
>>> tnet = TemporalNetwork(from_array=G)
Or for an already defined object:
>>> tnet.network_from_array(G)
Contact representation¶
The contact representation (see below) is a dictionary which a key called contacts includes a contact list of lists and some additional metadata. Here the argument is from_dict should be called.
>>> C = {'contacts': [[0,1,2],[1,0,0]],
'nettype': 'bu',
'netshape': (2,2,3),
't0': 0,
'nodelabels': ['A', 'B'],
'timeunit': 'seconds'}
>>> tnet = TemporalNetwork(from_dict=C)
Or alternatively:
>>> tnet = TemporalNetwork()
>>> tnet.network_from_dict(C)
Pandas data frame¶
Using a pandas data frame the data can also be imported. Here the required columns are: i, j and t (the first two are nodes, the latter is time index). The column weight is also needed for weighted networks.
>>> import pandas as pd
>>> netin = {'i': [0,0,1,1], 'j': [1,2,2,2], 't': [0,0,0,1], 'weight': [0.5,0.75,0.25,1]}
>>> df = pd.Data Frame(data=netin)
>>> tnet = TemporalNetwork(from_df=df)
>>> tnet.network
i j t weight
0 0 1 0 0.50
1 0 2 0 0.75
2 1 2 0 0.25
3 1 2 1 1.00
List of edges¶
Alternatively a list of lists can be given to TemporalNetwork, in such cases each sublist should follow the order [i,j,t,[weight]]. For example:
>>> edgelist = [[0,1,0,0.5], [0,1,1,0.75]]
>>> tnet = TemporalNetwork(from_edgelist=edgelist)
>>> tnet.network
i j t weight
0 0 1 0 0.50
1 0 1 1 0.75
This creates two edges between nodes 0 and 1 at two different time-points with two weights.
Array/snapshot representation¶
The array/snapshort representation is a three dimensional numpy array. The dimensions are (node,node,time).
The positives of arrays are that they is easy to understand and manipulate. The downside is that any meta-information about the network is lost and, when the networks are big, can use a lot of memory.
Contact representation¶
Note, the contact representation is going to be phased out in favour for the TemporalNetwork object with time.
The contact representations is a dictionary that can includes more information about the network than an array.
The keys in the dictionary include ‘contact’ (node,node,timestamp) which define all the edges in te network. A weights key is present in weighted networks containing the weights. Other keys for meta-information include: ‘dimord’ (dimension order), ‘Fs’ (sampling rate), ‘timeunit’, ‘nettype’ (if network is weighted/binary, undirected/directed), ‘timetype’, nodelabels (node labels), t0 (the first time point).
Converting between contact and graphlet representations¶
Converting between the two different network representations is quite easy. Let us generate a random network that consists of 3 nodes and 5 time points.
import teneto
import numpy as np
# For reproducibility
np.random.seed(2018)
# Number of nodes
N = 3
# Number of time-points
T = 5
# Probability of edge activation
p0to1 = 0.2
p1to1 = .9
G = teneto.generatenetwork.rand_binomial([N,N,T],[p0to1, p1to1],'graphlet','bu')
# Show shape of network
print(G.shape)
You can convert a graphlet representation to contact representation with: teneto.utils.graphlet2contact
C = teneto.utils.graphlet2contact(G)
print(C.keys)
To convert the opposite direction, type teneto.utils.contact2graphlet:
G2 = teneto.utils.contact2graphlet(C)
G==G2